-Search query
-Search result
Showing all 50 items for (author: sone & k)

PDB-8rx0: 
(NEDD8)-CRL2VHL-MZ1-Brd4BD2-Ub(G76S, K48C)-UBE2R1(C93K, S138C, C191S, C223S)-Ub
Method: single particle / : Crowe C, Nakasone MA
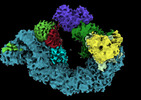
EMDB-19567: 
Closed crosslinked structure of (NEDD8)-CRL2VHL-MZ1-Brd4BD2-Ub(G76S, K48C)-UBE2R1(C93K, S138C, C191S, C223S)-Ub
Method: single particle / : Ciulli A, Crowe C, Nakasone MA
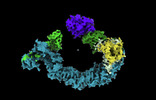
EMDB-19569: 
Open non-crosslinked structure Brd4BD2-MZ1-(NEDD8)-CRL2VHL
Method: single particle / : Ciulli A, Crowe C, Nakasone MA

EMDB-29298: 
The structure of a hibernating ribosome in the Lyme disease pathogen
Method: single particle / : Sharma MR, Manjari SR, Agrawal EK, Keshavan P, Koripella RK, Majumdar S, Marcinkiewicz AL, Lin YP, Agrawal RK, Banavali NK

EMDB-29304: 
The structure of a 50S ribosomal subunit in the Lyme disease pathogen Borreliella burgdorferi
Method: single particle / : Sharma MR, Manjari SR, Agrawal EK, Keshavan P, Koripella RK, Majumdar S, Marcinkiewicz AL, Lin YP, Agrawal RK, Banavali NK

PDB-8fmw: 
The structure of a hibernating ribosome in the Lyme disease pathogen
Method: single particle / : Sharma MR, Manjari SR, Agrawal EK, Keshavan P, Koripella RK, Majumdar S, Marcinkiewicz AL, Lin YP, Agrawal RK, Banavali NK

PDB-8fn2: 
The structure of a 50S ribosomal subunit in the Lyme disease pathogen Borreliella burgdorferi
Method: single particle / : Sharma MR, Manjari SR, Agrawal EK, Keshavan P, Koripella RK, Majumdar S, Marcinkiewicz AL, Lin YP, Agrawal RK, Banavali NK

EMDB-17172: 
Ternary structure of intramolecular bivalent glue degrader IBG1 bound to BRD4 and DCAF16:DDB1deltaBPB
Method: single particle / : Cowan AD, Sundaramoorthy R, Nakasone MA, Ciulli A

PDB-8ov6: 
Ternary structure of intramolecular bivalent glue degrader IBG1 bound to BRD4 and DCAF16:DDB1deltaBPB
Method: single particle / : Cowan AD, Sundaramoorthy R, Nakasone MA, Ciulli A

EMDB-29397: 
Structure of Mycobacterium smegmatis Rsh bound to a 70S translation initiation complex
Method: single particle / : Majumdar S, Sharma MR, Manjari SR, Banavali NK, Agrawal RK

PDB-8fr8: 
Structure of Mycobacterium smegmatis Rsh bound to a 70S translation initiation complex
Method: single particle / : Majumdar S, Sharma MR, Manjari SR, Banavali NK, Agrawal RK

EMDB-16226: 
Giardia Ribosome in PRE-T Classical State (C)
Method: single particle / : Majumdar S, Emmerich AG, Sanyal S

EMDB-16228: 
Giardia Ribosome in PRE-T Hybrid State (D1)
Method: single particle / : Majumdar S, Emmerich AG, Sanyal S

EMDB-16235: 
Giardia Ribosome in PRE-T Hybrid State (D2)
Method: single particle / : Majumdar S, Emmerich AG, Sanyal S

PDB-8bsj: 
Giardia Ribosome in PRE-T Classical State (C)
Method: single particle / : Majumdar S, Emmerich AG, Sanyal S

PDB-8btd: 
Giardia Ribosome in PRE-T Hybrid State (D1)
Method: single particle / : Majumdar S, Emmerich AG, Sanyal S

PDB-8btr: 
Giardia Ribosome in PRE-T Hybrid State (D2)
Method: single particle / : Majumdar S, Emmerich AG, Sanyal S

EMDB-16211: 
Giardia ribosome in POST-T state (A1)
Method: single particle / : Majumdar S, Emmerich AG, Sanyal S

EMDB-16222: 
Giardia ribosome in POST-T state, no E-site tRNA (A6)
Method: single particle / : Majumdar S, Emmerich AG, Sanyal S

EMDB-16225: 
Giardia ribosome chimeric hybrid-like GDP+Pi bound state (B1)
Method: single particle / : Majumdar S, Emmerich AG, Sanyal S

PDB-8br8: 
Giardia ribosome in POST-T state (A1)
Method: single particle / : Majumdar S, Emmerich AG, Sanyal S

PDB-8brm: 
Giardia ribosome in POST-T state, no E-site tRNA (A6)
Method: single particle / : Majumdar S, Emmerich AG, Sanyal S

PDB-8bsi: 
Giardia ribosome chimeric hybrid-like GDP+Pi bound state (B1)
Method: single particle / : Majumdar S, Emmerich AG, Sanyal S

EMDB-30440: 
Cryo-EM analysis of the nonribosomal peptide synthetase, FmoA3
Method: single particle / : Sone K, Harada A, Kawai S, Urano N, Adachi N, Moriya T, Kawasaki M, Katsuyama Y, Senda T, Ohnishi Y

EMDB-23211: 
Cryo-EM structure of human ACE2 receptor bound to protein encoded by vaccine candidate BNT162b1
Method: single particle / : Lees JA, Han S

EMDB-23215: 
Cryo-EM structure of protein encoded by vaccine candidate BNT162b2
Method: single particle / : Lees JA, Han S

PDB-7l7f: 
Cryo-EM structure of human ACE2 receptor bound to protein encoded by vaccine candidate BNT162b1
Method: single particle / : Lees JA, Han S

PDB-7l7k: 
Cryo-EM structure of protein encoded by vaccine candidate BNT162b2
Method: single particle / : Lees JA, Han S

EMDB-8166: 
Structure of the S. cerevisiae alpha-mannosidase 1
Method: single particle / : Schneider S, Kosinski J

EMDB-8167: 
Structure of S. cerevesiae mApe1 dodecamer
Method: single particle / : Sachse C, Bertipaglia C

PDB-5jm0: 
Structure of the S. cerevisiae alpha-mannosidase 1
Method: single particle / : Schneider S, Kosinski J, Jakobi AJ, Hagen WJH, Sachse C

PDB-5jm9: 
Structure of S. cerevesiae mApe1 dodecamer
Method: single particle / : Sachse C, Bertipaglia C

EMDB-2896: 
Cryo-EM structure of helical ANTH and ENTH tubules on PI(4,5)P2-containing membranes
Method: single particle / : Skruzny M, Desfosses A, Prinz S, Dodonova SO, Gieras A, Uetrecht C, Jakobi AJ, Abella M, Hagen WJH, Schulz J, Meijers R, Rybin V, Briggs JAG, Sachse C, Kaksonen M

EMDB-2897: 
Cryo-EM structure of helical ANTH and ENTH tubules on PI(4,5)P2-containing membranes
Method: single particle / : Skruzny M, Desfosses A, Prinz S, Dodonova SO, Gieras A, Uetrecht C, Jakobi AJ, Abella M, Hagen WJH, Schulz J, Meijers R, Rybin V, Briggs JAG, Sachse C, Kaksonen M

PDB-5ahv: 
Cryo-EM structure of helical ANTH and ENTH tubules on PI(4,5)P2-containing membranes
Method: single particle / : Skruzny M, Desfosses A, Prinz S, Dodonova SO, Gieras A, Uetrecht C, Jakobi AJ, Abella M, Hagen WJH, Schulz J, Meijers R, Rybin V, Briggs JAG, Sachse C, Kaksonen M

PDB-4bip: 
Homology model of coxsackievirus A7 (CAV7) full capsid proteins.
Method: single particle / : Seitsonen JJT, Shakeel S, Susi P, Pandurangan AP, Sinkovits RS, Hyvonen H, Laurinmaki P, Yla-Pelto J, Topf M, Hyypia T, Butcher SJ

PDB-4biq: 
Homology model of coxsackievirus A7 (CAV7) empty capsid proteins.
Method: single particle / : Seitsonen JJT, Shakeel S, Susi P, Pandurangan AP, Sinkovits RS, Hyvonen H, Laurinmaki P, Yla-Pelto J, Topf M, Hyypia T, Butcher SJ

EMDB-5512: 
Icosahedral reconstruction of filled coxsackievirus A9 capsid-integrin alpha v beta 6 complex
Method: single particle / : Shakeel S, Seitsonen JJT, Kajander T, Laurinmaki P, Hyypia T, Susi P, Butcher SJ

EMDB-5514: 
Icosahedral reconstruction of empty coxsackievirus A9 capsid-integrin alpha v beta 6 complex
Method: single particle / : Shakeel S, Seitsonen JJT, Kajander T, Laurinmaki P, Hyypia T, Susi P, Butcher SJ

EMDB-5515: 
Icosahedral reconstruction of filled coxsackievirus A9 capsid
Method: single particle / : Shakeel S, Seitsonen JJT, Kajander T, Laurinmaki P, Hyypia T, Susi P, Butcher SJ

EMDB-5516: 
Icosahedral reconstruction of empty coxsackievirus A9 capsid
Method: single particle / : Shakeel S, Seitsonen JJT, Kajander T, Laurinmaki P, Hyypia T, Susi P, Butcher SJ

EMDB-5517: 
Asymmetric reconstruction of filled-integrin alpha v beta 6 complex
Method: single particle / : Shakeel S, Seitsonen JJT, Kajander T, Laurinmaki P, Hyypia T, Susi P, Butcher SJ

EMDB-5519: 
Electron cryo-tomography of coxsackievirus A9-integrin alpha v beta 6 complex
Method: electron tomography / : Shakeel S, Seitsonen JJT, Kajander T, Laurinmaki P, Hyypia T, Susi P, Butcher SJ

PDB-3j2j: 
Empty coxsackievirus A9 capsid
Method: single particle / : Shakeel S, Seitsonen JJT, Kajander T, Laurinmaki P, Hyypia T, Susi P, Butcher SJ

EMDB-2027: 
Coxsackievirus A7 (CAV7) empty capsid reconstruction at 6.09 angstrom resolution.
Method: single particle / : Seitsonen JJT, Shakeel S, Susi P, Pandurangan AP, Sinkovits RS, Hyvonen H, Laurinmaki P, Yla-Pelto J, Topf M, Hyypia T, Butcher SJ

EMDB-2028: 
Coxsackievirus A7 (CAV7) full capsid reconstruction at 8.23 angstrom resolution.
Method: single particle / : Seitsonen JJT, Shakeel S, Susi P, Pandurangan AP, Sinkovits RS, Hyvonen H, Laurinmaki P, Yla-Pelto J, Topf M, Hyypia T, Butcher SJ

EMDB-1688: 
CryoEM reconstruction of human parechovirus 1 complexed with integrin alphaV-beta3
Method: single particle / : Seitsonen JJT, Susi P, Heikkila O, Laurinmaki P, Hyypia T, Butcher SJ

EMDB-1689: 
CryoEM reconstruction of human parechovirus 1 complexed with integrin alphaV-beta6
Method: single particle / : Seitsonen JJT, Susi P, Heikkila O, Laurinmaki P, Hyypia T, Butcher SJ

EMDB-1690: 
CryoEM reconstruction of human parechovirus 1
Method: single particle / : Seitsonen JJT, Susi P, Heikkila O, Laurinmaki P, Hyypia T, Butcher SJ

EMDB-1422: 
Structure and composition of the Shigella flexneri "needle complex", a part of its type III secreton.
Method: single particle / : Blocker AJ, Jouihri N, Larquet E, Gounon P, Ebel F, Parsot C, Sansonetti P, Allaoui A
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
